The structure

TIM is an enzyme that catalyzes the reversible isomerization of dihydroxyacetone phosphate (DHAP) to glyceraldehyde-3-phosphate (GAP). It is called isomerization because the reaction consists basically in moving the carbonyl group (=O) from C2 to C1. The role of TIM is crucial in the glycolysis pathway, which is one of the first chain of reactions involved in the oxidation of sugars, a metabolism that is common to almost every organism on earth (plants, humans, yeasts, bacteria…).
The enzyme is an α/β protein, its homodimer consists of 2 identical subunits of 257 amino acids. Even if each subunit contains the active site, the enzyme is functional only in its oligomeric form. This is may due to the interaction of residues between the two subunits that contribute to the activation of the complex.
In each unit, a particular structure called the TIM barrel has been identified. It is a type of super secondary structure found in many proteins, it consists of eight α-helices and eight parallel β-sheets alternating along the protein backbone. The hydrophobic residues of both the β-sheets and of the α-helices are facing on the interior of the barrel, creating a hydrophobic environment in the active site.
 |
| de novo TIM barrel design, it is possible to see the internal (β-sheets) and external (α-helices) barrel. Source: https://commons.wikimedia.org/wiki/File:STIM11.png |
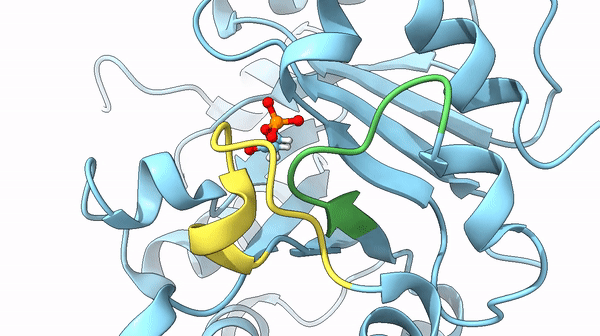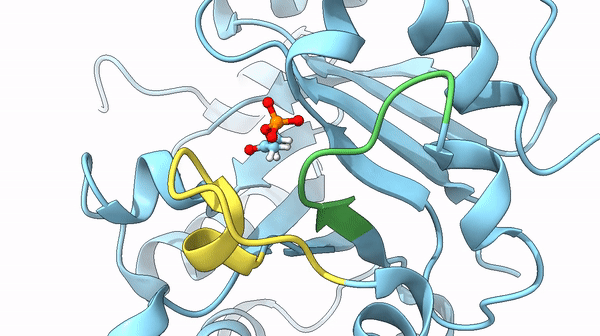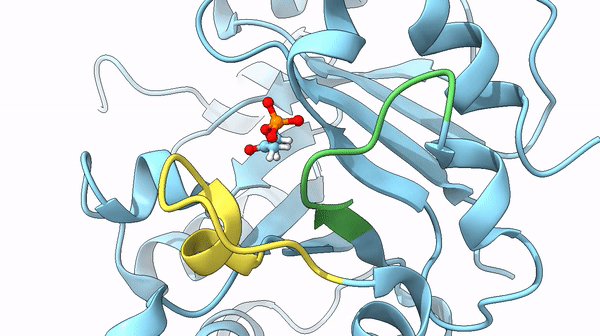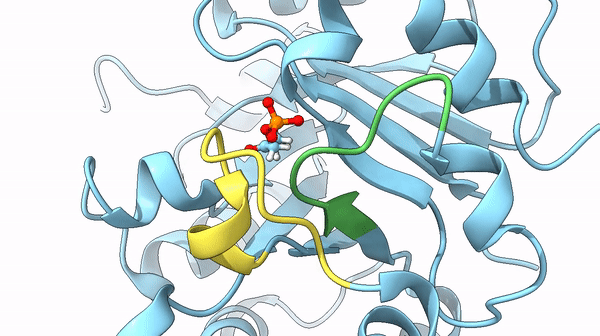
It was also found that a particular loop (6) acts as the lid on the active site, basically, after the active site bound the DHAP interacting with its phosphate dianion, it is trapped it in the internal pocket. Once the reaction is completed the lid opens and releases the substrate. The steps between the open and closed structure are complex, but they play a role also in the stabilization of particular intermediates of the reaction that could lead to undesired reactions otherwise.
 |
| The pattern of amino acid residue conservation among different taxa in TIM. Source: https://proteopedia.org/wiki/index.php/Triose_Phosphate_Isomerase |
The kinetics
The level
of efficiency of the enzyme is very high, so that, it has been tough to be “catalytically
perfect” because the rate of the reaction seems slowed down only by the
diffusion of substrate and product. With more recent studies it was found that
the reaction performed by TIM requires substantial conformational changes that
could limit the rate of the reaction. In particular, the previously cited movement of loops 6 and 7 may act as the limiting factor of the reaction
|
|
kcat, also known as Turnover Number (TN), it is a measure of the chemical conversion that a single enzyme can execute per second (s-1). The higher it is, the quicker is the reaction performed by the enzyme |
|
The Michaelis–Menten constant stands for the concentration of substrate necessary to reach ½ of the max velocity of the enzyme (M-1). The lower it is, the higher is the affinity between enzyme and substrate |
References
- Reaction
mechanism: Kulkarni, Y. S., Liao, Q., Byléhn,
F., Amyes, T. L., Richard, J. P., & Kamerlin, S. C. (2018). Role of
ligand-driven conformational changes in enzyme catalysis: modeling the
reactivity of the catalytic cage of triosephosphate isomerase. Journal
of the American Chemical Society, 140(11), 3854-3857.
- Structure of the loop: Liao, Q., Kulkarni, Y., Sengupta, U., Petrović, D., Mulholland, A. J., van der Kamp, M. W., ... & Kamerlin, S. C. L. (2018). Loop motion in triosephosphate isomerase is not a simple open and shut case. Journal of the American Chemical Society, 140(46), 15889-15903.
- Overview on the Michaelis-Menten kinetic: https://www.sciencedirect.com/topics/engineering/michaelis-menten-equation
- Take also a look at the metabolic interactive map: http://biochemical-pathways.com/#/map/1
- General information about TIM: https://proteopedia.org/wiki/index.php/Triose_Phosphate_Isomerase




0 Comments